Your new post is loading...
 Your new post is loading...
Authors: Tian Su, Ziwei Li, Yinghua Zhang, Junqiang Xu and Bin Xu.
Journal of the American Society for Horticultural Science (2024)
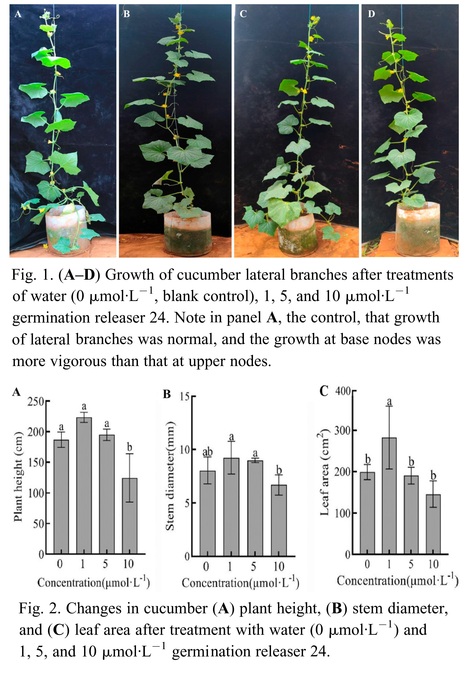
Abstract: "Cucumber (Cucumis sativus L.) belongs to the cucumber genus of the Cucurbitaceae family, and the selection of cultivars with minimal or no lateral branches can enhance the cultivation management efficiency. The growth of lateral branches is inhibited by strigolactone. To investigate the regulatory mechanism of strigolactone on the lateral branch development in cucumber, the cultivar LZ1 exhibiting multiple lateral branches was selected as the experimental material. The axillae of the plants were infiltrated with 1, 5, and 10 μmol·L−1 germination releaser 24 (GR24) at the four- to five-leaf stage. It was identified that 1 μmol·L−1 GR24 exhibited the most potent inhibitory effect on cucumber lateral branches. Additionally, exogenous strigolactone decreased the auxin content in the apical bud and axillae and increased the auxin content in the stem. This inhibited polar auxin transport in the axillary bud and promoted polar auxin transport in the apical bud. The content of strigolactone in the axilla region of cucumbers was elevated, whereas the synthesis and expression of cytokinin in the same area were reduced. A low concentration of GR24 induced the expression of cucumber branched 1 (csbrc1), whereas a high concentration of GR24 downregulated the expression of cucumber lateral suppressor (cscls) and blind (csblind), which inhibited the growth of cucumber lateral branches."
Authors: Nathalie Kuhn, Macarena Arellano, Claudio Ponce, Christian Hodar, Francisco Correa, Salvatore Multari, Stefan Martens, Esther Carrera, José Manuel Donoso and Lee A. Meisel
Journal of Plant Growth Regulation (2024)
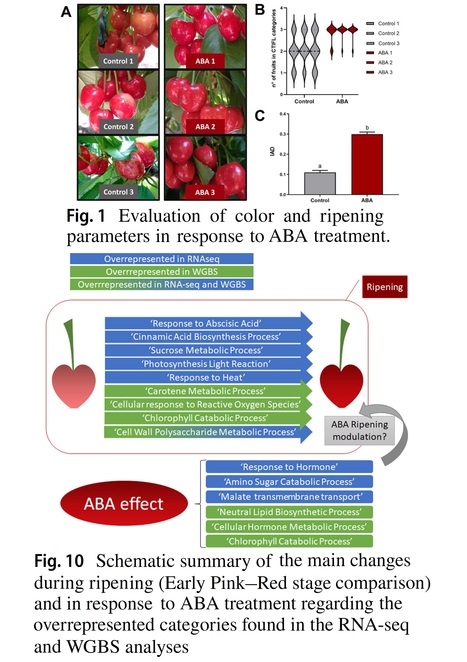
Abstract: "Abscisic acid (ABA) is a plant hormone that plays a key role in the ripening process of non-climacteric fruits, triggering pigment production, fruit softening, and sugar accumulation. Transcriptional studies show that ABA modifies the expression of several ripening-related genes, but epigenetic effects of ABA during this process are lacking. Therefore, this work aimed to perform transcriptomic and DNA methylation analyses of fruit samples treated with ABA during the fruit ripening process in the non-climacteric sweet cherry model. RNA-seq analyses revealed an overrepresentation of transcripts annotated in functional categories related to ABA response, secondary metabolism, and sugar synthesis during fruit ripening. In contrast, Whole Genome Bisulfite Sequencing (WGBS) analyses revealed DNA hypomethylation in the 5′UTR region of genes related to carotene catabolism. Transcriptional and epigenetic regulation of genes encoding xyloglucan enzymes, associated with cell wall modifications, were also detected. ABA treatment enhanced fruit color development and the accumulation of ripening markers, including carotenoids and several anthocyanins. Gene Ontology analysis in the RNA-seq of ABA-treated fruits revealed expression variations in genes encoding members of the Aux/IAA and ARF families. In the WGBS analysis, genes encoding enzymes for cytokinin biosynthesis had differential DNA methylation after the ABA treatment. Our work identified ABA-modulated factors at the genetic and epigenetic levels, suggesting complex hormone networks controlling non-climacteric sweet cherry fruit ripening."
Authors: Takuya Uragami, Takatoshi Kiba, Mikiko Kojima, Yumiko Takebayashi, Yuzuru Tozawa, Yuki Hayashi, Toshinori Kinoshita and Hitoshi Sakakibara.
bioRxiv (2024)
Abstract: "The directional and sequential flow of cytokinin in plants is organized by a complex network of transporters. Genes involved in several aspects of cytokinin transport have been characterized, but a large part of the elaborate system remains elusive. In this study, we have identified ABCC4 as a cytokinin efflux transporter gene. Using a transient expression system in tobacco leaves, we screened Arabidopsis transporter genes and isolated ATP-BINDING CASSETTE TRANSPORTER C4 (ABCC4). Further validation through drug-induced expression in Arabidopsis and heterologous expression in budding yeast revealed that ABCC4 effluxes the active form of cytokinins. During the seedling stage, ABCC4 was highly expressed in roots, and its expression was up-regulated in response to cytokinin application. Loss-of-function mutants of ABCC4 displayed enhanced primary root elongation, similar to mutants impaired in cytokinin biosynthesis or signaling, which was suppressed by exogenous trans-zeatin treatment. In contrast, overexpression of the gene led to suppression of root elongation. These results suggest that ABCC4 plays a role in the efflux of active cytokinin, thereby contributing to root growth regulation. Our findings contribute to unraveling the many complexities of cytokinin flow and enhance our understanding of the regulatory mechanisms underlying root system development in plants."
Authors: Yang Yu, Haixia Yu, Jing Peng, Wang Jinsong Yao, Yi Peng Wang, Feng Li Zhang, Shi Rong Wang, Yajie Zhao, Xiang Yu Zhao, Xian Sheng Zhang and Ying Hua Su.
Plant Communications (2024)
Editor's view: TaLAX1 can promote shoot regeneration, thereby boosting genetic transformation and genome editing in wheat by activating TaGRFs and TaGIF1, cytokinin biosynthesis genes, and auxin-response genes. Homologs of TaLAX1 can also enhance the regeneration capacity of maize and soybean.
Abstract: "In the realm of genetically transformed crops, the process of plant regeneration holds utmost significance. However, the low regeneration efficiency of several wheat varieties currently restricts the use of genetic transformation for gene functional analysis and improved crop production. This research explores overexpression of TaLAX PANICLE1 (TaLAX1), which markedly enhances regeneration efficiency, thereby boosting genetic transformation and genome editing in wheat. Particularly noteworthy is the substantial increase in regeneration efficiency of common wheat varieties previously regarded as recalcitrant to genetic transformation. Our study shows that increased expression of TaGROWTH-REGULATING FACTOR (TaGRF) genes, alongside that of their co-factor, TaGRF-INTERACTING FACTOR 1 (TaGIF1), enhances cytokinin accumulation and auxin response, which may play pivotal roles in the improved regeneration and transformation of TaLAX1-overexpressing wheat plants. Overexpression of TaLAX1 homologs also significantly increases the regeneration efficiency of maize and soybean, suggesting that both monocot and dicot crops can benefit from this enhancement. Our findings shed light on a gene that enhances wheat genetic transformation and elucidate molecular mechanisms that potentially underlie wheat regeneration."
Authors: Vojtěch Schmidt, Roman Skokan, Thomas Depaepe, Katarina Kurtović, Samuel Haluška, Stanislav Vosolsobě, Roberta Vaculíková, Anthony Pil, Petre Ivanov Dobrev, Václav Motyka, Dominique Van Der Straeten and Jan Petrášek.
Nature Communications (2024)
Editor's view: Here, the authors show that the biosynthesis of many compounds in green algae preceded their recruitment in phytohormone signaling and metabolism in land plants.
Abstract: "The genomes of charophyte green algae, close relatives of land plants, typically do not show signs of developmental regulation by phytohormones. However, scattered reports of endogenous phytohormone production in these organisms exist. We performed a comprehensive analysis of multiple phytohormones in Viridiplantae, focusing mainly on charophytes. We show that auxin, salicylic acid, ethylene and tRNA-derived cytokinins including cis-zeatin are found ubiquitously in Viridiplantae. By contrast, land plants but not green algae contain the trans-zeatin type cytokinins as well as auxin and cytokinin conjugates. Charophytes occasionally produce jasmonates and abscisic acid, whereas the latter is detected consistently in land plants. Several phytohormones are excreted into the culture medium, including auxin by charophytes and cytokinins and salicylic acid by Viridiplantae in general. We note that the conservation of phytohormone biosynthesis and signaling pathways known from angiosperms does not match the capacity for phytohormone biosynthesis in Viridiplantae. Our phylogenetically guided analysis of established algal cultures provides an important insight into phytohormone biosynthesis and metabolism across Streptophyta. Genomic evidence dates the origins of most phytohormones to terrestrialization or later."
Authors: Pengcheng Li, Zhihai Zhang, Gui Xiao, Zheng Zhao, Kunhui He, Xiaohong Yang, Qingchun Pan, Guohua Mi, Zhongtao Jia, Jianbing Yan, Fanjun Chen and Lixing Yuan.
Theoretical and Applied Genetics (2024)
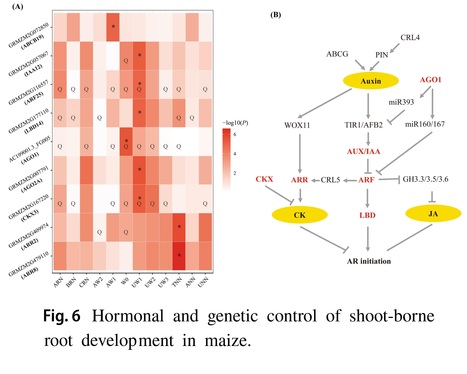
Abstract: "Efficient nutrient and water acquisition from soils depends on the root system architecture (RSA). However, the genetic determinants underlying RSA in maize remain largely unexplored. In this study, we conducted a comprehensive genetic analysis for 14 shoot-borne root traits using 513 inbred lines and 800 individuals from four recombinant inbred line (RIL) populations at the mature stage across multiple field trails. Our analysis revealed substantial phenotypic variation for these 14 root traits, with a total of 389 and 344 QTLs identified through genome-wide association analysis (GWAS) and linkage analysis, respectively. These QTLs collectively explained 32.2–65.0% and 23.7–63.4% of the trait variation within each population. Several a priori candidate genes involved in auxin and cytokinin signaling pathways, such as IAA26, ARF2, LBD37 and CKX3, were found to co-localize with these loci. In addition, a total of 69 transcription factors (TFs) from 27 TF families (MYB, NAC, bZIP, bHLH and WRKY) were found for shoot-borne root traits. A total of 19 genes including PIN3, LBD15, IAA32, IAA38 and ARR12 and 19 GWAS signals were overlapped with selective sweeps. Further, significant additive effects were found for root traits, and pyramiding the favorable alleles could enhance maize root development. These findings could contribute to understand the genetic basis of root development and evolution, and provided an important genetic resource for the genetic improvement of root traits in maize."
Via Jean-Michel Ané
Authors: Jiangzhe Zhao, Jingqi Wang, Jie Liu, Penghong Zhang, Guzel Kudoyarova, Chang-Jun Liu and Kewei Zhang.
Plant Communications (2024)
Abstract: "Cytokinins are a type of mobile phytohormone that regulate plant growth, development, and environmental adaptability. The major cytokinin species include isopentenyl adenine (iP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin (DZ). The spatial distributions of different cytokinin species in different organelles, cells, tissues, and organs are primarily shaped by biosynthesis via isopentenyltransferases (IPT), cytochrome P450 monooxygenase, and 5’-ribonucleotide phosphohydrolase, and by conjugation or catabolism via glycosyltransferase or cytokinin oxidase/dehydrogenase (CKX). Cytokinins bind to histidine receptor kinases (HKs) in the endoplasmic reticulum (ER) or plasma membrane (PM) and relay signals to response regulators (RRs) in the nucleus by shuttle proteins known as histidine phosphotransfer proteins (HPs). The movements of cytokinins from sites of biosynthesis to signal perception sites usually require long-distance, intercellular, and intracellular transport. In the past decade, ATP-binding cassette (ABC) transporters, purine permeases (PUP), AZA-GUANINE RESISTANT (AZG) transporters, equilibrative nucleoside transporters (ENT), and Sugars Will Eventually be Exported Transporters (SWEET) have been characterized as involved in cytokinin transport processes. This review begins by introducing the spatial distributions of various cytokinins and the subcellular localizations of the proteins involved in cytokinin metabolism and signaling. Highlights focus on an inventory of the characterized transporters involved in cytokinin compartmentalization, including long-distance, intercellular, and intracellular transport, and the regulation of spatial distributions of cytokinins by environmental cues. Future directions for cytokinin research are also discussed."
Authors: Dongmei Chen, Tianhui Liao, Wenjun Ye, Zhichao Jin and Shichao Ren.
Advanced Agrochem (2024)
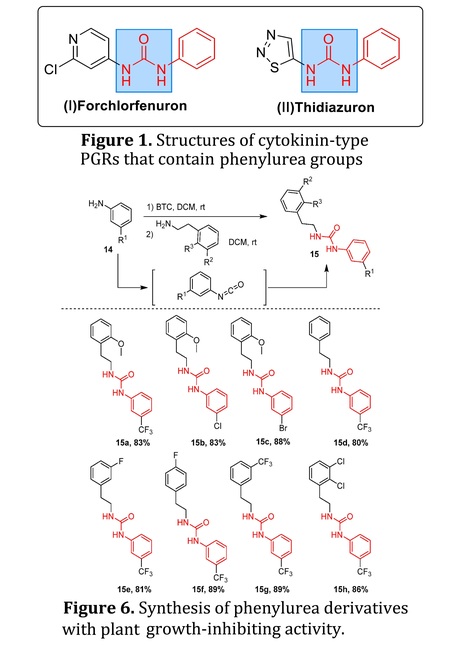
Abstract: "Plant growth regulators (PGRs) are chemical substances that imitate the functions of phytohormones to enhance the crop yield and the harvest process. Phenylurea-derived plant growth regulators are known for their excellent efficacy in promoting fruit growth, particularly in kiwifruit, grapes, and melons. Phenylurea derivatives represent one class of the highly efficient and versatile PGRs. Specifically, forchlorfenuron (CPPU, N-(2-chloro-4-pyridinyl)-N’-phenylurea) exhibits similar growth-regulating efficacy to cytokinins and has a significant impact on the plant growth and the crop yield. As a result, there is growing interest in exploring the incorporation of various phenylurea moieties into agrochemicals to enhance their regulatory properties on crops. This review aims to provide a comprehensive overview on representative synthetic approaches for phenylurea derived PGRs. Additionally, we provide our perspective on the future development in this active research field."
Authors: Yuwen Zhang, Xingliang Duan, Zhen Wang, Yuanda Lv, Weicong Qi, Lun Li, Le Luo and Wei Xuan.
Biochemical and Biophysical Research Communications (2024)
Highlights • Both synthetic AtCEPs and AtCEP5 overexpression suppress primary root elongation and lateral root formation. • The regulation of CEPs on root development requires their receptors CEPRs. • Molecular evidence reveals that CEPs inhibit root development by modulating auxin and cytokinin signaling.
Abstract: "C-terminally encoded peptides (CEPs) are peptide hormones that function as mobile signals coordinating crucial developmental programs in plants. Previous studies have revealed that CEPs exert negative regulation on root development through interaction with CEP receptors (CEPRs), CEPR DOWNSTREAMs (CEPDs), the cytokinin receptor ARABIDOPSIS HISTIDINE KINASE (AHKs) and the transcriptional repressor Auxin/Indole-3-Acetic Acid (AUX/IAA). However, the precise molecular mechanisms underlying CEPs-mediated regulation of root development via auxin and cytokinin signaling pathways still necessitate further detailed investigation. In this study, we examined prior research and elucidated the underlying molecular mechanisms. The results showed that both synthetic AtCEPs and overexpression of AtCEP5 markedly suppressed primary root elongation and lateral root (LR) formation in Arabidopsis. Molecular biology and genetics elucidated how CEPs inhibit root growth by suppressing auxin signaling while promoting cytokinin signaling. In summary, this study elucidated the inhibitory effects of AtCEPs on Arabidopsis root growth and provided insights into their potential molecular mechanisms, thus enhancing our comprehension of CEP-mediated regulation of plant growth and development."
Authors: Grace A. Johnston, Hannah M. Berry, Mikiko Kojima, Hitoshi Sakakibara and Cristiana T. Argueso.
bioRxiv (2024)
Abstract: "Plant immunity activation often results in suppression of plant growth, particularly in the case of constitutive immune activation. We discovered that signaling of the phytohormone cytokinin (CK), known to regulate plant growth through the control of cell division and shoot apical meristem (SAM) activity, can be suppressed by negative crosstalk with the defense phytohormones jasmonic acid (JA), and most evidently, salicylic acid (SA). We show that changing the negative crosstalk of SA on CK signaling in autoimmunity mutants by targeted increase of endogenous CK levels results in plants resistant to pathogens from diverse lifestyles, and relieves suppression of reproductive growth. Moreover, such changes in crosstalk result in a novel reproductive growth phenotype, suggesting a role for defense phytohormones in the SAM, likely through regulation of nitrogen response and cellular redox status. Our data suggest that targeted phytohormone crosstalk engineering can be used to achieve increased reproductive growth and pathogen resistance."
Authors: Nabila El Arbi, Ann-Kathrin Schürholz, Marlene U. Handl , Alexei Schiffner, Inés Hidalgo Prados, Liese Schnurbusch, Christian Wenzl, Xin’Ai Zhao, Jian Zeng, Jan U. Lohmann and Sebastian Wolf.
EMBO Journal (2024)
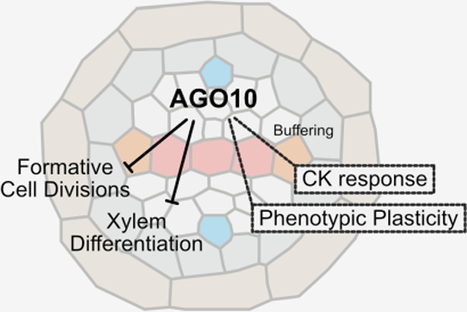
Synopsis: In the Arabidopsis root vascular tissue, a gradient of miRNA165/6 controls the abundance of HD-ZIP III transcription factors, which in turn control cell fate and spatially restrict vascular cell proliferation to specific cells. This work shows that vascular development requires ARGONAUTE10 to sequester miRNA165/6, thus protecting HD-ZIP III transcripts from degradation. • AGO10 controls formative cell divisions in the centre of the root stele, with its loss leading to ectopic xylem strands, increased procambial cell number, and enhanced root growth. • Consistent with a role in protecting HD-ZIP III transcripts from miRNA165/6-mediated degradation, AGO10 is mostly expressed in the centre of the meristematic stele. • AGO10 promotes HD-ZIP III activity to suppress cytokinin responses in the stele. • AGO10 mutants outperform the wild type under water-limiting conditions. • However, AGO10 is required for phenotypic robustness upon reduced carbon availability.
Abstract: A key question in plant biology is how oriented cell divisions are integrated with patterning mechanisms to generate organs with adequate cell type allocation. In the root vasculature, a gradient of miRNA165/6 controls the abundance of HD-ZIP III transcription factors, which in turn control cell fate and spatially restrict vascular cell proliferation to specific cells. Here, we show that vascular development requires the presence of ARGONAUTE10, which is thought to sequester miRNA165/6 and protect HD-ZIP III transcripts from degradation. Our results suggest that the miR165/6-AGO10-HDZIP III module acts by buffering cytokinin responses and restricting xylem differentiation. Mutants of AGO10 show faster growth rates and strongly enhanced survival under severe drought conditions. However, this superior performance is offset by markedly increased variation and phenotypic plasticity in sub-optimal carbon supply conditions. Thus, AGO10 is required for the control of formative cell division and coordination of robust cell fate specification of the vasculature, while altering its expression provides a means to adjust phenotypic plasticity.
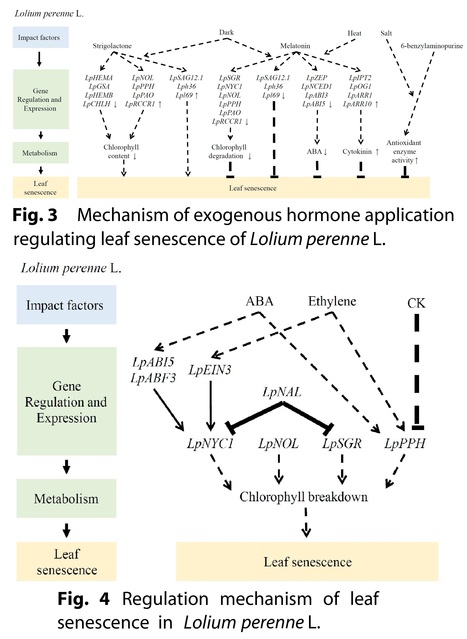
Authors: Kangning Zhang, Hongli Xie, Jiangqi Wen, Jing Zhang, Zeng-Yu Wang, Bin Xu and Maofeng Chai.
Grass Research (2024)
Abstract: "Leaf senescence is a complex biological process regulated by development, phytohormones, and various environmental factors. For forage and turf grasses, controlling leaf senescence can greatly improve forage quality, the amenity of lawn and turf, and the grasses’ stress tolerances. Leaf senescence involves a multitude of gene regulation and metabolic changes, including the alteration of chlorophyll metabolism. Here, we summarized the recent progress of studies on leaf senescence in major forage and turf grass species, such as Medicago truncatula, M. sativa, Lolium perenne, Panicum virgatum, and Agrostis stolonifera, to provide an insight into the development of effective methods for delaying leaf senescence in grass species."
Authors: Pramod Rathor, Punita Upadhyay, Aman Ullah, Linda Yuya Gorim and Malinda S. Thilakarathna.
AoB Plants (2024)
Abstract: "Humic acids have been widely used for centuries to enhance plant growth and productivity. The beneficial effects of humic acids have been attributed to different functional groups and phytohormone-like compounds enclosed in macrostructure. However, the mechanisms underlying the plant growth-promoting effects of humic acids are only partially understood. We hypothesize that the bio-stimulatory effect of humic acids is mainly due to the modulation of innate pathways of auxin and cytokinin biosynthesis in treated plants. A physiological investigation along with molecular characterization was carried out to understand the mechanism of bio-stimulatory effects of humic acid. A gene expression analysis was performed for the genes involved in auxin and cytokinin biosynthesis pathways in wheat seedlings. Furthermore, Arabidopsis thaliana transgenic lines generated by fusing the auxin-responsive DR5 and cytokinin-responsive ARR5 promoter to ß-glucuronidase (GUS) reporter were used to study the GUS expression analysis in humic acid treated seedlings. This study demonstrates that humic acid treatment improved the shoot and root growth of wheat seedlings. The expression of several genes involved in auxin (Tryptophan Aminotransferase of Arabidopsis and Gretchen Hagen 3.2) and cytokinin (Lonely Guy3) biosynthesis pathways was up-regulated in humic acid treated seedlings compared to the control. Furthermore, GUS expression analysis showed that bioactive compounds of humic acid stimulate endogenous auxin and cytokinin-like activities. This study is the first report in which using ARR5 :GUS lines we demonstrate the biostimulants activity of humic acid."
|
Authors: Yuqi Liu, Shangyu Chen, Sikander Pal, Jingquan Yu, Yanhong Zhou, Lam-Son Phan Tran and Xiaojian Xia.
New Crops (2024)
Abstract: "Plants have evolved varied structures for environmental adaptation. Shoot branching, as a part of plant architecture, influences the allocation of sugars produced by photosynthesis and thus greatly impacts crop yields. The activity of axillary meristem, and apical dominance governs the shoot branching patterns. In this review, we summarize the key factors involved in the formation of lateral branches, and the mechanisms of how these factors are interconnected. In particular, we focus on recent advances in understanding how sugar and environmental signals affect the hormonal signaling network to regulate apical dominance. Ultimately, we propose that epigenetic modifications are critical mechanisms underlying the plasticity of shoot branching, and that precise targeted gene editing is promising for shaping the ideal plant architecture."
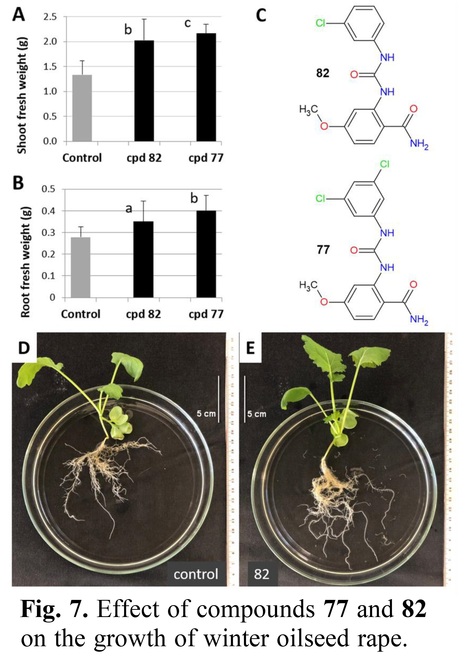
Authors: Jaroslav Nisler, Pavel Klimeš, Radka Končitíková, Alena Kadlecová, Jiří Voller, Mahfam Chalaki, Michael Karampelias, Nino Murvanidze, Stefaan P O Werbrouck, David Kopečný, Libor Havlíček, Nuria de Diego, Pierre Briozzo, Solange Moréra, David Zalabák and Lukáš Spíchal.
Journal of Experimental Botany (2024)
Abstract: "Cytokinin oxidase/dehydrogenase (CKX) inhibitors reduce the degradation of cytokinins in plants and thereby may improve the efficiency of agriculture and plant tissue culture-based practices. Here, we report a synthesis and structure-activity relationship study of novel urea derivatives concerning their CKX inhibitory activity. The best compounds showed sub-nanomolar IC50 values with maize ZmCKX1, the lowest value yet documented. Other CKX isoforms of maize (Zea mays) and Arabidopsis were also inhibited very effectively. The binding mode of four compounds was characterized based on high-resolution crystal complex structures. Using the soil nematode Caenorhabditis elegans, and human skin fibroblasts, key CKX inhibitors with low toxicity were identified. These compounds enhanced the shoot regeneration of Lobelia, Drosera, and Plectranthus, as well as the growth of Arabidopsis and Brassica napus. At the same time, a key compound (namely 82), activated a cytokinin primary response gene ARR5:GUS and cytokinin sensor TCSv2:GUS, without activating the Arabidopsis cytokinin receptors AHK3 and AHK4. This strongly implies that the effect of compound 82 is due to the upregulation of cytokinin signalling. Overall, this work presents highly effective and easily prepared CKX inhibitors with a low risk of environmental toxicity for further investigation of their potential in agriculture and biotechnology."
Authors: Kien Huu Nguyen, Zihan Li, Chengliang Wang, Chien Van Ha, Cuong Duy Tran, Mostafa Abdelrahman, Hoi Xuan Pham, Khuat Huu Trung, Tran Dang Khanh, Ha Duc Chu, Mohammad Golam Mostofa, Yasuko Watanabe, Yaping Wang, Yuchen Miao, Keiichi Mochida, Sikander Pal, Weiqiang Li and Lam-Son Phan Tran.
Plant Stress (2024)
Highlights: • Antagonistic interaction of CK and MAX2 signaling in plant adaption to drought. • max2 mutant is more drought-sensitive, while ahk2 ahk3 double mutant is more drought-tolerant. • AHK-MAX2 interaction affects ABA response, ROS balance and leaf hydration under drought. • Trait marker gene expression patterns align with drought tolerance hierarchy of investigated mutants
Abstract: "Understanding the mechanisms, especially those associated with phytohormones, of plant drought adaptation is crucial for sustaining agricultural production in the era of climate change. Arabidopsis histidine kinases (AHKs), an integral part of the cytokinin signaling pathway, and more axillary growth 2 (MAX2), a key component of the strigolactone and karrikin signaling pathways are reported to act as negative and positive regulators, respectively, in plant adaption to drought. However, the potential interaction between these signaling pathways in plant drought adaptation is not fully understood. To address this query, we assessed drought tolerance levels and associated phenotypic and physiological traits of the max2 single mutant, ahk2 ahk3 double mutant, ahk2 ahk3 max2 triple mutant, and wild-type (WT) Arabidopsis thaliana plants. Our findings revealed a distinct hierarchy in drought tolerance among these genotypes, as indicated by the differences in plant growth and stress survival rates. Specifically, the max2 mutant displayed the lowest drought tolerance level, followed by WT, ahk2 ahk3 max2, and ahk2 ahk3 plants. Additionally, the observed changes in leaf relative water content, leaf surface temperature, and cuticle formation were coherently aligned with the observed hierarchy of drought tolerance levels. Under drought conditions, the max2 mutant exhibited higher oxidative stress and membrane damage, as evidenced by increased levels of reactive oxygen species (ROS), malondialdehyde, and electrolyte leakage. In contrast, the ahk2 ahk3 and ahk2 ahk3 max2 mutants showed low and intermediate levels, respectively, for these parameters. The max2 mutant displayed reduced sensitivity, whereas ahk2 ahk3 and ahk2 ahk3 max2 mutants demonstrated high and intermediate sensitivities, respectively, to exogenous abscisic acid (ABA) treatments. Additionally, the expression analysis of several genes associated with the investigated drought tolerance-related traits showed a positive correlation between the transcript levels and corresponding trait(s) in both mutant and WT plants under drought conditions. Our results collectively indicate the presence of an antagonistic interaction between AHK and MAX2 signaling pathways in plant drought adaptation, impacting ABA responsiveness, leaf water retention, cuticle development, and ROS homeostasis. Findings of this study provide a valuable foundation for developing agricultural methods to improve plant drought resilience."
Authors: Jorge Hernández-García, Antonio Serrano-Mislata, María Lozano-Quiles, Cristina Úrbez, María A. Nohales, Noel Blanco-Touriñán, Huadong Peng, Rodrigo Ledesma-Amaro and Miguel A. Blázquez.
PNAS (2024)
Significance: DELLA proteins are plant-specific transcriptional hubs integrating environmental signals with endogenous cues. In order to regulate downstream processes, DELLAs modulate the activity of hundreds of transcription factors (TFs) and transcriptional regulators in various ways. Here, we describe the molecular mechanism underlying DELLA coactivator function. We show that DELLAs act as transcriptional activators by interacting with the Mediator complex subunit MED15. This interaction is necessary to regulate a specific subset of DELLA-dependent responses that are mediated by transcriptional coactivation, but not those regulated by TF sequestration. We further show that this mechanism is present in bryophyte DELLAs and thus represents a conserved mechanism of DELLA function in land plants.
Abstract: "DELLA proteins are negative regulators of the gibberellin response pathway in angiosperms, acting as central hubs that interact with hundreds of transcription factors (TFs) and regulators to modulate their activities. While the mechanism of TF sequestration by DELLAs to prevent DNA binding to downstream targets has been extensively documented, the mechanism that allows them to act as coactivators remains to be understood. Here, we demonstrate that DELLAs directly recruit the Mediator complex to specific loci in Arabidopsis, facilitating transcription. This recruitment involves DELLA amino-terminal domain and the conserved MED15 KIX domain. Accordingly, partial loss of MED15 function mainly disrupted processes known to rely on DELLA coactivation capacity, including cytokinin-dependent regulation of meristem function and skotomorphogenic response, gibberellin metabolism feedback, and flavonol production. We have also found that the single DELLA protein in the liverwort Marchantia polymorpha is capable of recruiting MpMED15 subunits, contributing to transcriptional coactivation. The conservation of Mediator-dependent transcriptional coactivation by DELLA between Arabidopsis and Marchantia implies that this mechanism is intrinsic to the emergence of DELLA in the last common ancestor of land plants."
Authors: Lei Liang and Xiangyang Hu.
Phyton (2024)
Abstract: "With the rapid development of modern molecular biology and bioinformatics, many studies have proved that transcription factors play an important role in regulating the growth and development of plants. SPATULA (SPT) belongs to the bHLH transcription family and participates in many processes of regulating plant growth and development. This review systemically summarizes the multiple roles of SPT in plant growth, development, and stress response, including seed germination, flowering, leaf size, carpel development, and root elongation, which is helpful for us to better understand the functions of SPT."
Authors: Pragya Yadav, Shashank Kumar Yadav, Meenu Singh, Madan Pal Singh and Viswanathan Chinnusamy.
Plant Physiology Reports (2024)
Abstract: "Rice (Oryza sativa L.) is one of the key food crops in ensuring global food security and accounts for nearly one fourth of the world’s total food supply. Rice production is significantly affected by abiotic stresses. Cytokinins (CKs) play a pivotal role in the regulation of the plant growth, development and abiotic stress responses. In present study, genome-wise analysis led to the identification of 10 Isopentenyl transferase (IPT) gene family members, encoding rate-limiting enzymes in CK biosynthesis pathway in rice. Phylogenetic analysis categorized IPTs into four subfamilies that are structurally and functionally conserved across species with similar gene structures, motifs, domains, cis-acting elements were identified. We performed a genome-wide identification of IPT family members from the rice genome, identified their chromosomal location, gene structure, protein properties, cis-element and phylogeny. Further, Real-time PCR analysis was carried out to understand the spatio-temporal and stress responsive expression of IPT gene family members. OsIPT1, OsIPT2, OsIPT4, OsIPT7, OsIPT8 and OsIPT10 showed similar expression level in both root and shoot of seedling or more expression in root, while OsIPT3, OsIPT5, OsIPT6 and OsIPT9 showed higher expression in shoot as compared with root in seedlings. Abiotic stress responsive expression analysis showed that abiotic stresses reduced the expression of all family members except IPT6. Analysis of expression of IPTs in rice cv. Kitaake (low biomass and low yielding) and Taipei (high biomass and high yielding) showed that expression of IPTs were higher in different tissues in Taipei. Present study has also elaborately described the tissue and stage specific expression and for further analysis of IPT in the regulation of biotic and abiotic stress resistance, growth, and development in rice, and provides a comprehensive analysis of the OsIPT gene family, including structures and functions."
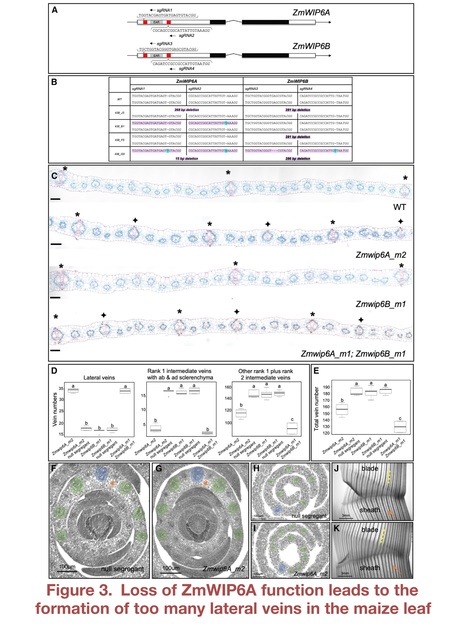
Authors: Daniela Vlad, Maricris Zaidem, Chiara Perico, Olga Sedelnikova, Samik Bhattacharya and Jane A. Langdale.
Current Biology (2024)
Editor's view: Vlad et al. discover a conserved mechanism that determines whether procambial stem cells in grass leaf primordia will develop into large lateral or smaller intermediate veins. The mechanism is revealed through the analysis of mutants defective in the activity of a zinc finger transcription factor of the WIP6 family that is named TOO MANY LATERALS.
Highlights • Function of WIP6 ortholog TML is conserved in maize and rice • TML is expressed in procambial cells during early leaf vein development • tml mutants develop large lateral veins in places where smaller veins should form • TML function suppresses lateral vein development in a subset of procambial initials
Abstract: "Grass leaves are invariantly strap shaped with an elongated distal blade and a proximal sheath that wraps around the stem. Underpinning this shape is a scaffold of leaf veins, most of which extend in parallel along the proximo-distal leaf axis. Differences between species are apparent both in the vein types that develop and in the distance between veins across the medio-lateral leaf axis. A prominent engineering goal is to increase vein density in leaves of C3 photosynthesizing species to facilitate the introduction of the more efficient C4 pathway. Here, we discover that the WIP6 transcription factor TOO MANY LATERALS (TML) specifies vein rank in both maize (C4) and rice (C3). Loss-of-function tml mutations cause large lateral veins to develop in positions normally occupied by smaller intermediate veins, and TML transcript localization in wild-type leaves is consistent with a role in suppressing lateral vein development in procambial cells that form intermediate veins. Attempts to manipulate TML function in rice were unsuccessful because transgene expression was silenced, suggesting that precise TML expression is essential for shoot viability. This finding may reflect the need to prevent the inappropriate activation of downstream targets or, given that transcriptome analysis revealed altered cytokinin and auxin signaling profiles in maize tml mutants, the need to prevent local or general hormonal imbalances. Importantly, rice tml mutants display an increased occupancy of veins in the leaf, providing a step toward an anatomical chassis for C4 engineering. Collectively, a conserved mechanism of vein rank specification in grass leaves has been revealed.
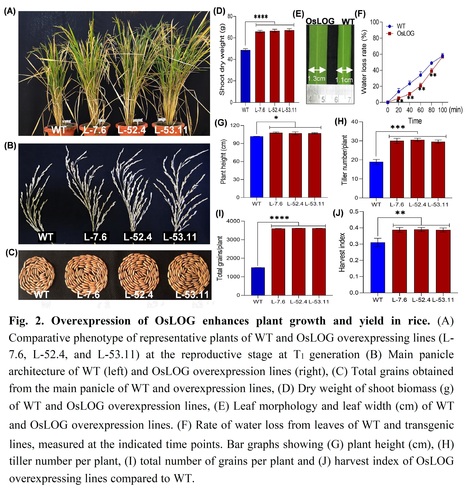
Authors: Ray Singh Rathore, Manjari Mishra, Ashwani Pareek and Sneh Lata Singla-Pareek.
Plant Physiology and Biochemistry (2024)
Highlights: • Overexpression of Lonely Guy (OsLOG) gene in rice leads to increase in the active form (trans-zeatin) of cytokinin. • OsLOG mediated elevated cytokinin levels promote ideal plant architecture and rice yield. • Elevated cytokinin levels in OsLOG transgenic lines enhance tolerance to drought and salinity stress and reduce yield penalty through maintenance of redox homeostasis.
Abstract: "Meristem activity is important for normal plant growth as well as adaptive plastic development under abiotic stresses. Cytokinin has been recognized to have a major role in regulating meristem function which is controlled by cytokinin activating enzymes by fine-tuning the concentrations and spatial distribution of its bioactive forms. It was previously reported that LONELY GUY (LOG) acts in the direct activation pathway of cytokinin in rice shoot meristems. LOG has a cytokinin specific phosphoribohydrolase activity, which transforms inactive cytokinin nucleotides into active free bases. Here, we explored the role of OsLOG in controlling meristem activity mediated by cytokinin and its effects on growth, development, and stress resilience of rice plants. Overexpression of OsLOG in rice led to significant alterations in cytokinin levels in the inflorescence meristem, leading to enhanced plant growth, biomass and grain yield under both non-stress as well as stress conditions such as drought and salinity. Moreover, our study provides insight into how overexpression of OsLOG improves the ability of plants to withstand stress. The OsLOG-overexpressing lines exhibit reduced accumulation of H2O2 along with elevated antioxidant enzyme activities, thereby maintaining better redox homeostasis under stress conditions. This ultimately reduces the negative impact of stresses on grain yield and improves harvest index, as evidenced by observations in the OsLOG-overexpressing lines. In summary, our study emphasizes the diverse role of OsLOG, not only in regulating plant growth and yield via cytokinin but also in enhancing adaptability to abiotic stresses. This highlights its potential to improve crop yield and promote sustainable agriculture."
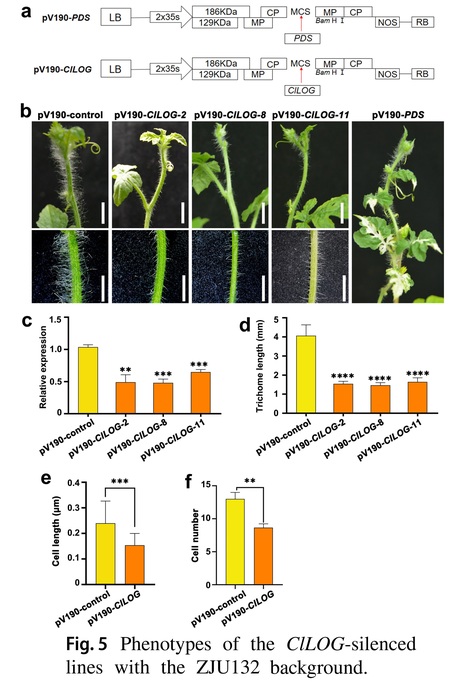
Authors: Yuyuan Ma, Yu Wang, Zhiqin Zhou, Runqin Zhang, Yiru Xie, Yihan Zhang, Yongming Bo, Xiaolong Lyu, Jinghua Yang, Mingfang Zhang and Zhongyuan Hu.
Theoretical and Applied Genetics (2024)
Key message: The ClLOG gene encoding a cytokinin riboside 5ʹ-monophosphate phosphoribohydrolase determines trichome length in watermelon, which is associated with its promoter variations.
Abstract: "Trichomes, which are differentiated from epidermal cells, are special accessory structures that cover the above-ground organs of plants and possibly contribute to biotic and abiotic stress resistance. Here, a bulked segregant analysis (BSA) of an F2 population with significant variations in trichome length was undertaken. A 1.84-Mb candidate region on chromosome 10 was associated with trichome length. Resequencing and fine-mapping analyses indicated that a 12-kb structural variation in the promoter of Cla97C10G203450 (ClLOG) led to a significant expression difference in this gene in watermelon lines with different trichome lengths. In addition, a virus-induced gene silencing analysis confirmed that ClLOG positively regulated trichome elongation. These findings provide new information and identify a potential target gene for controlling multicellular trichome elongation in watermelon."
Authors: Wenzhe Yu, Li Luo, Xiangyu Qi, Yuman Cao, Jie An, Zhiguo Xie, Tianming Hu and Peizhi Yang.
Journal of Agricultural and Food Chemistry (2024)
Abstract: "Plant growth-promoting rhizobacteria have been shown to enhance plant tolerance to drought stress through various mechanisms. However, there is limited research on improving drought resistance in alfalfa by genetically modifying PGPR to produce increased levels of cytokinins. Herein, we employed synthetic biology approaches to engineer two novel strains of Sinorhizobium meliloti capable of overproducing trans-Zeatin and investigated their potential in enhancing drought tolerance in alfalfa. Our results demonstrate that alfalfa plants inoculated with these engineered S. meliloti strains exhibited reduced wilting and yellowing while maintaining higher relative water content under drought conditions. The engineered S. meliloti-induced tZ activated the activity of antioxidant enzymes and the accumulation of osmolytes. Additionally, the increased endogenous tZ content in plants alleviated the impact of drought stress on the alfalfa photosynthetic rate. However, under nondrought conditions, inoculation with the engineered S. meliloti strains had no significant effect on alfalfa biomass and nodule formation."
Authors: Chenyu Rong, Renren Zhang, Yuexin Liu, Zhongyuan Chang, Ziyu Liu, Yanfeng Ding and Chengqiang Ding.
Plant Cell Reports (2024)
Key message: Purine permease PUP11 is essential for rice seed development, regulates the seed setting rate, and influences the cytokinin content, sugar transport, and starch biosynthesis during grain development.
Abstract: "The distribution of cytokinins in plant tissues determines plant growth and development and is regulated by several cytokinin transporters, including purine permease (PUP). Thirteen PUP genes have been identified within the rice genome; however, the functions of most of these genes remain poorly understood. We found that pup11 mutants showed extremely low seed setting rates and a unique filled seed distribution. Moreover, seed formation arrest in these mutants was associated with the disappearance of accumulated starch 10 days after flowering. PUP11 has two major transcripts with different expression patterns and subcellular locations, and further studies revealed that they have redundant positive roles in regulating the seed setting rate. We also found that type-A Response Regulator (RR) genes were upregulated in the developing grains of the pup11 mutant compared with those in the wild type. The results also showed that PUP11 altered the expression of several sucrose transporters and significantly upregulated certain starch biosynthesis genes. In summary, our results indicate that PUP11 influences the rice seed setting rate by regulating sucrose transport and starch accumulation during grain filling. This research provides new insights into the relationship between cytokinins and seed development, which may help improve cereal yield."
Authors: Yafan Han, Minghao Qu, Zhongchi Liu and Chunying Kang.
The Plant Cell (2024)
One-sentence summary: An MYB transcription factor directly regulates cytokinin homeostasis to repress axillary bud growth and consequently affect crown formation in woodland strawberry.
Abstract: "Shoot branching affects plant architecture. In strawberry (Fragaria L.), short branches (crowns) develop from dormant axillary buds to form inflorescences and flowers. While this developmental transition contributes greatly to perenniality and yield in strawberry, its regulatory mechanism remains unclear and understudied. In the woodland strawberry (Fragaria vesca), we identified and characterized two independent mutants showing more crowns. Both mutant alleles reside in FveMYB117a, a R2R3-MYB transcription factor gene highly expressed in shoot apical meristems, axillary buds and young leaves. Transcriptome analysis revealed that the expression of several cytokinin pathway genes was altered in the fvemyb117a mutant. Consistently, active cytokinins were significantly increased in the axillary buds of the fvemyb117a mutant. Exogenous application of cytokinin enhanced crown outgrowth in the wild type, whereas the cytokinin inhibitors suppressed crown outgrowth in the fvemyb117a mutant. FveMYB117a binds directly to the promoters of the cytokinin homeostasis genes FveIPT2 encoding an isopentenyltransferase and FveCKX1 encoding a cytokinin oxidase to regulate their expression. Conversely, the type-B Arabidopsis response regulators FveARR1 and FveARR2b can directly inhibit the expression of FveMYB117a, indicative of a negative feedback regulation. In conclusion, we identified FveMYB117a as a key repressor of crown outgrowth by inhibiting cytokinin accumulation and provide a mechanistic basis for bud fate transition in an herbaceous perennial plant."
|


 Your new post is loading...
Your new post is loading...